Multiple CDM datasets can be aggregated into a single, logical dataset. This is done with the aggregation NcML element. There are several types of aggregation:
Download aggUnionSimple.ncml, cldc.mean.nc and lflx.mean.nc and place them together in a single directory. In the ToolsUI NcML Tab, open aggUnionSimple.ncml and examine it:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2"> <attribute name="title" type="string" value="Union cldc and lflx"/>
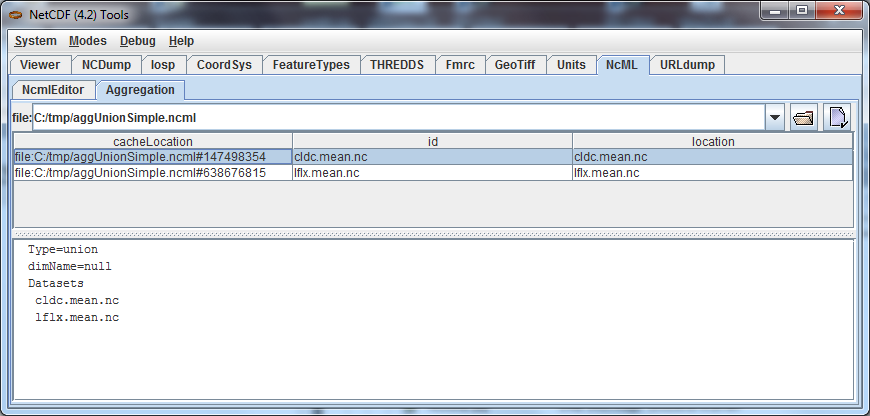
<aggregation type="union"> <netcdf location="cldc.mean.nc"/>
<netcdf location="lflx.mean.nc"/> </aggregation> </netcdf>
This NcML constructs a dataset by creating the union of two netCDF files (note there is no location attribute in the outer netcdf element).
Go to the NcML/Aggregation Tab of ToolsUI and cut and paste the full path of the aggUnionSimple.ncml file:

The files that comprise the aggregation are listed in the top table, various information about the aggregation is
listed in the lower pane. Select one of the files, and right click to bring up the context menu. Choose "Open
as NetCDF File" so that you can look at the contents of that file. You can open the nested files in the Viewer
NCDump ![]() . They look like the following (details
omitted):
. They look like the following (details
omitted):
netcdf cldc.mean.nc {
dimensions:
time = UNLIMITED; // (456 currently)
lat = 21;
lon = 360;
variables:
float lat(lat=21);
float lon(lon=360);
double time(time=456);
short cldc(time=456, lat=21, lon=360);
}
netcdf lflx.mean.nc {
dimensions:
time = UNLIMITED; // (456 currently)
lat = 21;
lon = 360;
variables:
float lat(lat=21);
float lon(lon=360);
double time(time=456);
short lflx(time=456, lat=21, lon=360);
}
In the ToolsUI Viewer Tab, open aggUnionSimple.ncml and examine it:
netcdf aggUnionSimple.xml {
dimensions:
time = UNLIMITED; // (456 currently)
lat = 21;
lon = 360;
variables:
float lat(lat=21);
float lon(lon=360);
double time(time=456);
short cldc(time=456, lat=21, lon=360);
short lflx(time=456, lat=21, lon=360);
}
The cldc variable will be read from cldc.mean.nc, while the lflx variable will be read from lflx.mean.nc. All the other variables, dimensions, and attributes are taken from the first file that declares them.
A Union dataset is constructed by transferring objects (dimensions, attributes, groups, and variables) from the nested datasets in the order the nested datasets are listed. If an object with the same name already exists, it is skipped. You need to pay close attention to dimensions and coordinate variables, which must match exactly across nested files.
Download aggExisting.ncml, jan.nc and feb.nc and place them together in a single directory. In the ToolsUI NcML Tab, open aggExisting.ncml and examine it:
This NcML constructs a dataset by declaring "time" the aggregation dimension. Any variable that has that dimension as its outer dimension is an aggregation variable. The dimension must be the outer (slowest varying) dimension. There must be an existing coordinate variable named time.
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinExisting"> <netcdf location="jan.nc" /> <netcdf location="feb.nc" /> </aggregation>
</netcdf>
Go to the NcML/Aggregation Tab of ToolsUI and cut and paste the full path of the aggExisting.ncml file:
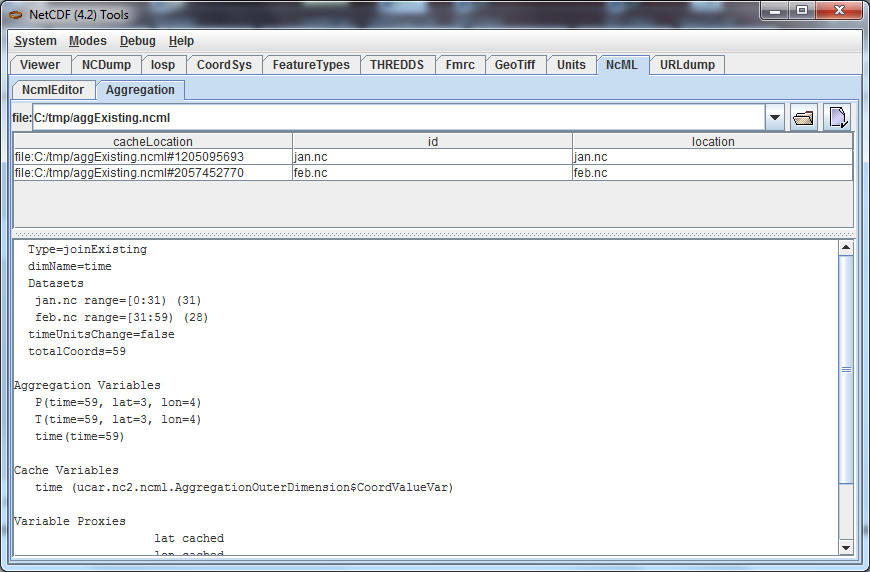
In the lower pane, you can see the list of variables that will be aggregated (P,T,time) and that the first 31 indices come from jan.nc, and the next 28 from feb.nc.
Open the nested files in the ToolsUI NCDump Tab. They look like the following (details omitted):
netcdf jan.nc {
dimensions:
lat = 3;
lon = 4;
time = 31;
variables:
double P(time=31, lat=3, lon=4);
double T(time=31, lat=3, lon=4);
float lat(lat=3);
float lon(lon=4);
int time(time=31);
}
netcdf feb.nc {
dimensions:
lat = 3;
lon = 4;
time = 28;
variables:
double P(time=28, lat=3, lon=4);
double T(time=28, lat=3, lon=4);
float lat(lat=3);
float lon(lon=4);
int time(time=28);
}
In the ToolsUI Viewer Tab, open aggExisting.ncml and examine it using the NCDumpData icon located on the toolbar:
netcdf aggExisting.xml {
dimensions:
lat = 3;
lon = 4;
time = 59;
variables:
double P(time=59, lat=3, lon=4);
double T(time=59, lat=3, lon=4);
float lat(lat=3);
float lon(lon=4);
int time(time=59);
}
The variables P, T, and time are aggregation variables, because they have the aggregation dimension as their first (outermost) dimension. The first 31 data values are taken from the file jan.nc and the next 28 data values are taken from feb.nc. All the other variables, dimensions, and attributes are taken from the first file that declares them.
A JoinExisting dataset is constructed by transferring objects (dimensions, attributes, groups, and variables) from the nested datasets in the order the nested datasets are listed. All variables that use the aggregation dimension as their outer dimension are logically concatenated, in the order of the nested datasets. Variables that don't use the aggregation dimension are treated as in a Union dataset, i.e. skipped if one with that name already exists.
When the library opens the above NcML dataset, it has to read through all nested datasets, in order to find out the length of the time dimension. For large aggregations, this can be slow. In the example below, we have added the optional ncoords attribute on the nested datasets. In this case, only one dataset has to be opened immediately, and the others as needed for a data read request.
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinExisting"> <netcdf location="file:/test/temperature/jan.nc" ncoords="31"/> <netcdf location="file:/test/temperature/feb.nc" ncoords="28"/> </aggregation>
</netcdf>
Typically the coordinates for a JoinExisting aggregation are taken from the existing coordinate variable, as in the above example. If the coordinate is missing, you must define it in the NcML:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
(1)<variable name="time" shape="time" type="int">
<attribute name="units" value="days since 2000-01-01"/>
<attribute name="_CoordinateAxisType" value="Time" />
(2) <values start="0" increment="1" />
</variable>
<aggregation dimName="time" type="joinExisting">
<netcdf location="file:/test/temperature/jan.nc" ncoords="31"/>
<netcdf location="file:/test/temperature/feb.nc" ncoords="28"/>
</aggregation>
</netcdf>
These are the ways that coordinate values may be assigned to a JoinExisting coordinate:
<aggregation dimName="time" type="joinExisting"> <netcdf location="file:/test/temperature/janAvgWeek.nc" coordValue="1038 7823 12983 43400"/> <netcdf location="file:/test/temperature/febAvgWeek.nc" coordValue="66234 89237 108736 123494"/> </aggregation>
<aggregation dimName="time" type="joinExisting" timeUnitsChange="true"> <netcdf location="file:/test/temperature/janAvgWeek.nc" /> <netcdf location="file:/test/temperature/febAvgWeek.nc" /> </aggregation>The timeUnitsChange feature requires that all the aggregation coordinate values will be read in when the dataset is opened.
The previous example "joined" variables along their existing outer dimension. Another common case is to aggregate variables by creating a new outer dimension. Each existing variable becomes one "slice" of the compound variable (a slice holds the index of one dimension constant, e.g. humidity(3,:,:,:)). The following NcML joins variables from three separate files into a single variable, by creating a new dimension of length 3:
Download aggNew.ncml, time0.nc, time1.nc and time2.nc and place them together in a single directory. In the ToolsUI NcML Tab, open aggNew.ncml and examine it:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
(1)<aggregation dimName="time" type="joinNew">
(2) <variableAgg name="T"/>
(3) <netcdf location="time0.nc" coordValue="0"/>
<netcdf location="time1.nc" coordValue="10"/>
<netcdf location="time2.nc" coordValue="99"/>
</aggregation>
</netcdf>
Go to the NcML/Aggregation Tab of ToolsUI and cut and paste the full path of the aggNew.ncml file:
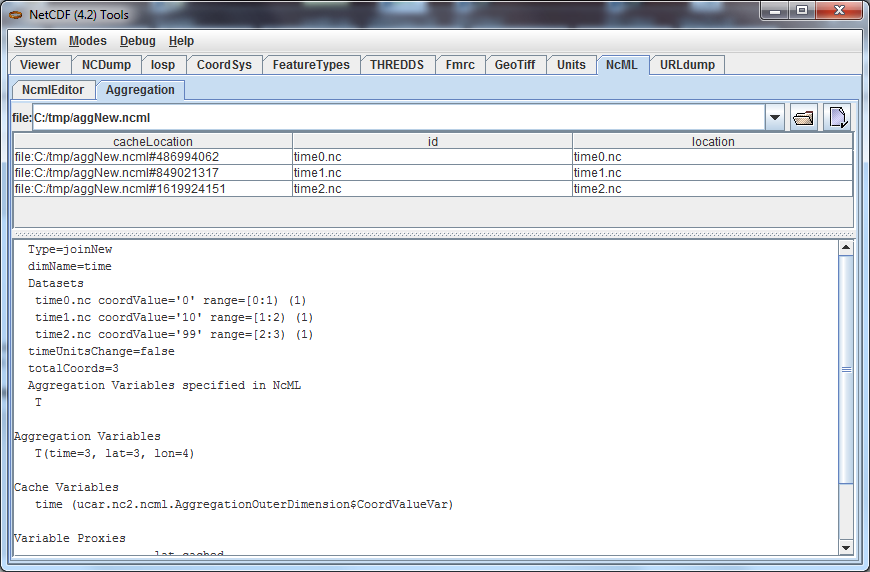
Open the nested files in the ToolsUI NCDump Tab. They all look like the following (details omitted):
netcdf time0.nc {
dimensions:
lat = 3;
lon = 4;
variables:
double T(lat=3, lon=4);
float lat(lat=3);
float lon(lon=4);
}
This will create the following dataset:
netcdf file:C:/dev/tds/thredds/docs/web/netcdf-java/ncml/examples/aggNew.ncml {
dimensions:
lat = 3;
lon = 4;
time = 3;
variables:
float lat(lat=3);
float lon(lon=4);
int time(time=3);
double T(time=3, lat=3, lon=4);
data:
time = {0, 10, 99}
}
So a new dimension and coordinate variable time(time) has been added, and the aggregation variable T now has time as its outer dimension. The data for T from the nested files are logically concatenated together.
A JoinNew dataset is constructed by transferring objects (dimensions, attributes, groups, and variables) from the nested datasets in the order the nested datasets are listed. All variables that are listed as aggregation variables are logically concatenated along the new dimension, in the order of the nested datasets. A coordinate Variable is created for the new dimension. Non-aggregation variables are treated as in a Union dataset, i.e. skipped if one of that name already exists.
A JoinNew aggregation has to create a new coordinate variable. In the above example, one was automatically created with type double, to match the coordValues specified on the netcdf elements. However, it has no units or other attributes. To specify attributes on the coordinate system, you can use the following (download aggNewCoord.ncm):
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
(1)<variable name="time" type="int" >
<attribute name="units" value="months since 2000-6-16 6:00"/>
<attribute name="_CoordinateAxisType" value="Time" />
<values>0 1 2</values>
</variable>
(2)<aggregation dimName="time" type="joinNew">
<variableAgg name="T"/>
<netcdf location="time0.nc" />
<netcdf location="time1.nc" />
<netcdf location="time2.nc" />
</aggregation>
</netcdf>
Its not obvious from the NcML, but the aggregation element (2) is processed first, so that all of the objects of the aggregated datasets are available to be modified by other NcML elements, for example by (1).
This will create the following dataset:
netcdf file:C:/dev/tds/thredds/docs/web/netcdf-java/ncml/examples/aggNew.ncml {
dimensions:
lat = 3;
lon = 4;
time = 3;
variables:
float lat(lat=3);
float lon(lon=4);
int time(time=3;)
:units = "months since 2000-6-16 6:00";
:_CoordinateAxisType = "Time";
double T(time=3, lat=3, lon=4);
data:
time = {0, 1, 2}
}
There are several ways that coordinate values are assigned to a JoinNew coordinate:
Note that you must explicitly specify the coordinate variable in order to assign attributes to it, which is something you are likely to need to do, for example defining a units attribute is usually necessary. Assigning the _CoordinateAxisType type is one way to make sure that the Coordinate layer correctly identifies the coordinate type. Using CF Conventions is strongly recommended.
Also note that, contrary to previous versions of NcML aggregation, you do not need to define a dimension element for the aggregation dimension (e.g. <dimension name="time"> and must not use the old form <dimension name="time" length="0" /> as it will override the dimension created by the aggregation.
For all aggregations, the aggregation element is processed first, so that the objects (dimensions, attributes, groups, and variables) from the nested datasets exist and can be modified by other NcML elements.
Its often convenient to use all the files in some directory without having to name them individually. The following example scans all of the files in the directory /data/model (and its subdirectories) which end in ".nc". By default, the files are ordered by sorting on the filename.
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinExisting">
<scan location="/data/model/" suffix=".nc" />
</aggregation>
</netcdf>
When opening a joinExisting aggregation using a scan element, each matching file must be opened in order to determine its size. This can be slow if there are a large number of files. In the case where you specify the files individually, you could add the ncoords attribute for speed. In the THREDDS Data Server, the information is cached, so that subsequent requests do not need to open each file until data is requested. However, see the section on caching.
A joinNew type aggregation does not incur this expense, since there is always exactly one step per file:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinNew">
<variableAgg name="T"/>
<scan location="/data/goes/" suffix=".gini" />
</aggregation>
</netcdf>
In a joinNew aggregation, the problem is how to assign coordinate values to each step? If you do nothing, a String-valued coordinate variable will be defined, whose values are the filenames. Better is to specify the coordinate variable and assign it values:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<variable name="time" type="int" shape="time" >
<attribute name="units" value="hours since 2000-01-01 00:00"/>
<attribute name="_CoordinateAxisType" value="Time" />
<values start="0" increment="1" />
</variable>
<aggregation dimName="time" type="joinNew">
<variableAgg name="T"/>
<scan location="/data/goes/" suffix=".gini" />
</aggregation>
</netcdf>
You can also explicitly list the values:
<values>12.0 27.0 39.0 51.0</values>
If the values are evenly spaced, you can use the start/increment form, and you don't need to know the number of files:
<values start="12.0" increment="25.0"/>
For the common case that the filename contains date information from which you can derive a time coordinate, you can
use the dateFormatMark attribute (download aggDateFormat.ncml
and cg.zip, unzip the latter and place in your data directory). In the ToolsUI
NcML Tab, open aggDateFormat.ncml and change the scan location to point to your
data directory, and then save it ![]() :
:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinExisting" recheckEvery="4 sec">
<scan location="CHANGE THIS" dateFormatMark="CG#yyyyDDD_HHmmss" suffix=".nc" subdirs="false" />
</aggregation>
</netcdf>
The dateFormatMark attribute is used on joinNew aggregation, as well as joinExisting if there is only one time slice in each file, to create date coordinate values out of the filename. It consists of a section of text, a '#' marking character, then a java.text.SimpleDateFormat string. The number of characters before the # is skipped in the filename, then the next part of the filename must match the SimpleDateFormat string, then it ignores any trailing text. For example:
Filename: CG2006158_120000h_usfc.nc DateFormatMark: CG#yyyyDDD_HHmmss
The net effect is to add a coordinate variable, whose values are ISO 8601 formatted date/time Strings, with a _CoordinateAxisType of "Time", so the example NcML will show this (details skipped):
netcdf file:C:/dev/tds/thredds/cdm/src/test/data/ncml/aggExistingOne.xml {
dimensions:
altitude = 1;
lat = 29;
lon = 26;
time = 3;
variables:
float altitude(altitude=1);
float lat(lat=29);
float lon(lon=26);
float CGusfc(time=3, altitude=1, lat=29, lon=26);
String time(time=3);
:_CoordinateAxisType = "Time";
:long_name = "time coordinate";
:standard_name = "time";
data:
time = "2006-06-07T12:00:00Z", "2006-06-07T13:00:00Z", "2006-06-07T14:00:00Z"
}
The scan element allows you to specify that all of the files in a directory (and its
subdirectories, with an optional suffix filter) are included in the aggregation. The files are sorted alphabetically
on the filename, unless you specify a dateFormatMark attribute, in which case they are sorted by
the Date derived from the filename, which is also used for the coordinate values.
When you use a scan element to define a collection of files, the case where the set of files may change as new files are added or deleted requires special attention.
There are situations where you need to indicate how often the directories should be rescanned.
You indicate how often the directories should be rescanned using the recheckEvery attribute:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinNew" recheckEvery="15 min" >
<variableAgg name="T"/>
<scan location="/data/goes/" suffix=".gini" />
</aggregation>
</netcdf>
The value of recheckEvery must be a udunit time unit, e.g. uses units of sec, min, hour, day, etc. If you do not specify a recheckEvery attribute, the collection will be assumed to be non-changing.
When using the scan element on directories whose contents may change, you must use a recheckEvery attribute. It specifies the maximum time before changes will be detected by a newly opened NcML dataset. An existing NcML dataset will not notice the changes, and you can get FileNotFoundException if the component files are deleted.
Note that the recheckEvery attribute specifies how out-of-date you are willing to allow your changing datasets to be, not how often the data changes. If you want updates to be seen within 5 min, use 5 minutes here, regardless of the frequency of updating.
For large collection of files, one wants to avoid opening every single file each time the dataset is accessed. Instead we only want to open the files that are actually needed to fulfill a data request. Generally this is straightforward, except for discovering the number and values of the aggregation coordinate variable for type joinExisting. This is because we have to know the size of the aggregation dimension when we open the dataset, even before we read any data. For practical purposes, we often need to know the coordinate values immediately also.
To help solve this problem, you should enable Aggregation Caching in your application, by telling the ucar.nc2.ncml.Aggregation class where it can cache information, by calling the static method (see javadoc for more details):
// Enable Aggregation caching. Every hour, delete stuff older than 30 days
Aggregation.setPersistenceCache( new DiskCache2("/.unidata/aggCache", true, 60 * 24 * 30, 60));
When this is enabled, joinExisting aggregations will save information to special XML files in the specified directory, in order to avoid opening every file to obtain its coordinate values, each time the dataset is opened. Instead, the first time it is opened, the values are read, then subsequent opens will use the cached values.
If using a scan element on changing directories, be sure to specify the recheckEvery attribute to make sure that the cached information gets updated.
One can nest netcdf elements in aggregation, for example:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2">
<aggregation dimName="time" type="joinExisting">
<netcdf>
<aggregation type="union">
<netcdf location="file:C:/test/path/temperature_20080101.nc" />
<netcdf location="file:C:/test/path/salinity_20080101.nc" />
</aggregation>
</netcdf>
<netcdf>
<aggregation type="union">
<netcdf location="file:C:/test/path/temperature_20080102.nc" />
<netcdf location="file:C:/test/path/salinity_20080102.nc" />
</aggregation>
</netcdf>
</aggregation>
</netcdf>
See also: Annotated NcML Schema
 This document is maintained by Unidata. Send comments to THREDDS support. Last updated: May 2011
This document is maintained by Unidata. Send comments to THREDDS support. Last updated: May 2011