A NetcdfFile provides read-only access to datasets through the netCDF API (to write data, use NetcdfFileWriteable). Use the static NetcdfFile.open methods to open a netCDF file, an HDF5 file, or any other file which has an IOServiceProvider implementation that can read the file with the NetCDF API. Use NetcdfDataset.open for more general reading capabilities, including OPeNDAP, NcML, and THREDDS datasets.
A simple way to open a NetcdfFile:
String filename = "C:/data/my/file.nc"; NetcdfFile ncfile = null;
try {
ncfile = NetcdfFile.open(filename);
process( ncfile);
} catch (IOException ioe) {
log("trying to open " + filename, ioe); } finally { if (null != ncfile) try { ncfile.close(); } catch (IOException ioe) { log("trying to close " + filename, ioe); } }
There is some boilerplate overhead in handling possible IOExceptions and ensuring that the file is properly closed, but these are very important when creating robust applications.
The NetcdfFile class will open local files for which an IOServiceProvider implementation exists. The current set of files that can be opened by the CDM are here.
When you open any of these files, the IOSP populates the NetcdfFile with a set of Variable, Dimension, Attribute, and possibly Group, Structure, and EnumTypedef objects that describe what data is available for reading from the file. These objects are called the structural metadata of the dataset, and they are read into memory at the time the file is opened. The data itself is not read until requested.
If NetcdfFile.open is given a filename that ends with ".Z", ".zip", ".gzip", ".gz", or ".bz2", it will uncompress the file before
opening, preferably in the same directory as the original file. See DiskCache
for more details.
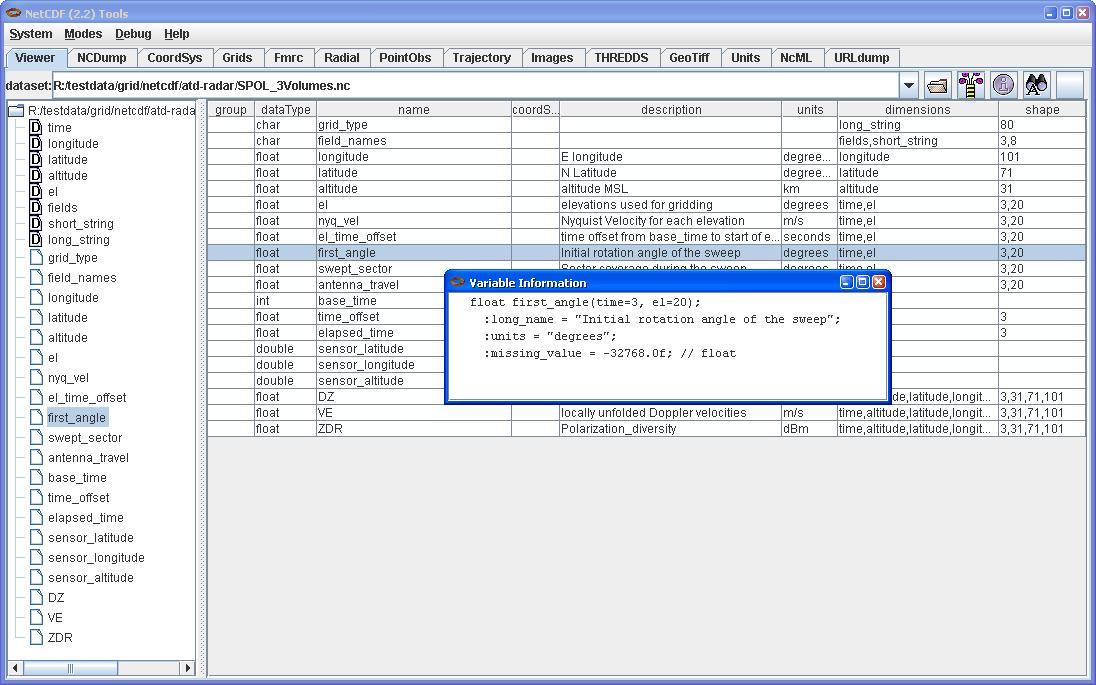
The NetCDF Tools User Interface (aka ToolsUI) is a program for browsing and debugging NetCDF files. You can start it from the command line, or launch it from webstart.
In this screen shot, the Viewer tab is shown displaying a NetCDF file in a tree view (on the left) and a table view of the Variables (on the right). By selecting a Variable, right clicking to get the context menu, and choosing Show Declaration, you can also display the variable's declaration in CDL in a popup window. The NCDump Data option from the same context menu will allow you to dump all or part of a Variable's data values from a window like this:
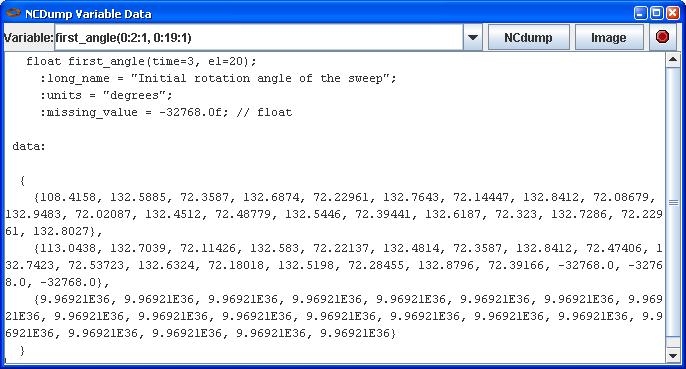
Note that you can edit the Variable's ranges (first_angle(0:2:1, 0:19:1) in this example) to dump just a subset of the data. These are
expressed
with Fortran 90 array section syntax, using zero-based indexing. For example, varName(
12:22
, 0:100:2, :, 17) specifies an array section for a four dimensional variable. The first dimension includes all the elements from 12 to 22 inclusive, the second dimension includes the elements from 0 to 100 inclusive with a stride of 2, the third includes all the elements in that dimension, and the fourth includes just the 18th element.
Here's the equivilent code to dump the data from your program:
String varName = "first_angle";
Variable v = ncfile.findVariable(varName); if (null == v) return;
try {
Array data = v.read("0:2:1, 0:19:1");
NCdumpW.printArray(data, varName, System.out, null);
} catch (IOException ioe) {
log("trying to read " + varName, ioe); } catch (InvalidRangeException e) { log("invalid Range for " + varName, e); }
If you want all the data in a variable, use:
Array data = v.read();
When you want to subset the data, you have a number of options, all of which have situations where they are the most convenient. Assume you have a 3D variable:
short temperature(time=10, lat=500, lon=720);
and you want to extract the third time step, and all lat and lon points, then use:
int[] origin = new int[] {2, 0, 0};
int[] size =
new int[] {1, 500, 720};
Array data3D = v.read(origin, size);
Array data2D = data3D.reduce();
Notice that the result of reading a 3D Variable is a 3D Array. To make it a 2D array call Array.reduce(), which removes any dimensions of length 1.
Or suppose you want to loop over all time steps, and make it general to handle any sized 3 dimensional variable:
int[] varShape = v.getShape();
int[] origin = new int[3];
int[] size = new int[] {1, varShape[1], varShape[2]};
for (int i = 0; i < varShape[0]; i++) {
origin[0] = i;
Array data2D = v.read(origin, size).reduce(0);
}
In this case, we call reduce(0), to reduce dimension 0, which we know has length one, but leave the other two dimensions alone.
Note that varShape holds the total number of elements that can be read from the variable; origin is the starting index, and size is the number of elements to read. This is different from the Fortran 90 array syntax, which uses the starting and ending array indices (inclusive):
Array data = v.read("2,0:499,0:719");
If you want strided access, you can use the Fortran 90 string routine:
Array data = v.read("2,0:499:2,0:719:2");
For general programing, use the read method that takes a List of ucar.ma2.Range, which follows the Fortran 90 array syntax, taking the starting and ending indices (inclusive), and an optional stride:
List ranges = new ArrayList();
ranges.add(new Range(2,2));
ranges.add(new Range(0,499,2));
ranges.add(new Range(0,719,2));
Array data2 = v.read(ranges).reduce();
For example, to loop over all time steps with any sized 3 dimensional variable, taking every second lat and every second lon point.
List ranges = new ArrayList();
ranges.add(null);
ranges.add(new Range(0, varShape[1]-1, 2));
ranges.add(new Range(0, varShape[2]-1, 2));
for (int i = 0; i < 3; i++) {
ranges.set(0, new Range(i, i));
Array data2D = v.read(ranges).reduce(0);
}
There are a number of useful static methods in the Range class for moving between Lists of Ranges and origin, size arrays:
int[] varShape = v.getShape(); //make the equivilent List of Ranges
List ranges = new ArrayList();
for (int i = 0; i < varShape.length; i++)
Range.appendShape( ranges, varShape[i]); // make the equivilent origin, size arrays
int[] origins = Range.getOrigin( ranges); int[] shapes = Range.getShapes( ranges);
There are convenience routines in the Variable class for reading scalar variables, for example:
double dval = v.readScalarDouble(); float fval = v.readScalarFloat(); int ival = v.readScalarInt(); ...
for example, the readScalarDouble() will read a scalar variable's single value as a double, converting it to double if needed. This can also be used for 1D variables with dimension length = 1, eg:
double height_above_ground(level=1);
These scalar routines are available in these data types: byte, double, float, int, long, short, and String. The String scalar method:
String sval = v.readScalarString();
can be used on scalar String or char, or 1D char variables of any size, such as:
char varname(name_strlen=77);
Once you read the data in, you usually have an Array object to work with. The shape of the Array will match the shape of the variable, if the entire data was read, or the shape of the section, if a subset was read in. There are a number of ways to access the data in the Array. Heres an example where you know you have a 2D array, and you want to keep track of what index you are on:
Array data = v.read(); int[] shape = data.getShape();
Index index = data.getIndex();
for (i=0; i<shape[0]; i++) { for (j=0; j<shape[1]; j++) { double dval = data.getDouble(index.set(i,j)); } }
If you want to iterate over all the data in a variable of any rank, without keeping track of the indices:
Array data = v.read();
double sum = 0.0;
IndexIterator ii = data.getIndexIterator();
while (ii.hasNext())
sum += ii.getDoubleNext();
You can also just iterate over a section of the data defined by a List of Ranges. The following iterates over every 5th point of each dimensions in an Array of arbitrary rank:
Array data = v.read(); int[] dataShape = data.getShape();
List ranges = new ArrayList();
for (int i = 0; i < dataShape.length; i++)
ranges.add( new Range(0, dataShape[i]-1, 5)); double sum = 0.0; IndexIterator ii = data.getRangeIterator(ranges);
while (ii.hasNext())
sum += ii.getDoubleNext();
In these examples, the data will be converted to double if needed.
If you know the Array's rank and type, you can cast to the appropriate subclass and use the get() and set() methods, for example:
ArrayDouble.D2 data = (ArrayDouble.D2) v.read(); int[] shape = data.getShape();
for (i=0; i<shape[0]; i++) { for (j=0; j<shape[1]; j++) { double dval = data.get(i,j); } }
There are a number of index reordering methods that operate on an Array, and return another Array with the same backing data storage, but with the indices modified in various ways:
public Array flip( int dim); // invert dimension public Array permute( int[] dims); // permute dimensions public Array reduce(); // rank reduction for any dims of length 1 public Array reduce(int dim); // rank reduction for specific dimension public Array section( Range[] ranges); // create logical subset public Array sectionNoReduce( Range[] ranges); // no rank reduction public Array slice(int dim, int val); // rank-1 subset public Array transpose( int dim1, int dim2); // transpose dimensions
The backing data storage for an Array is a 1D Java array of the corresponding type (double[] for ArrayDouble, etc) with length Array.getSize(). You can work directly with the Java array by extracting it from the Array, for example:
float[] javaArray = (float []) data.get1DJavaArray( float.class);
If the Array has the same type as the request, and the indices have not been reordered, this will return the backing array, otherwise it will return a copy with the requested type and correct index ordering.
There are a number of places where the library needs to write files to disk. If you end up using the file more than once, its useful to cache these files.
Before it writes the temporary file, it looks to see if it already exists. By default, it prefers to place the temporary file in the same directory as the original file. If it does not have write permission in that directory, by default it will use the directory ${user_home}/.unidata/cache/. You can change the directory by calling ucar.nc2.util.DiskCache.setDirectory().
You might want to always write temporary files to the cache directory, in order to manage them in a central place. To do so, call ucar.nc2.util.DiskCache.setCachePolicy( boolean alwaysInCache) with parameter alwaysInCache = true. You may want to limit the amount of space the disk cache uses (unless you always have data in writeable directories, so that the disk cache is never used). To scour the cache, call DiskCache.cleanCache(). For long running appplication, you might want to do this periodically in a background timer thread, as in the following example.
1) Calendar c = Calendar.getInstance(); // contains current startup time
c.add( Calendar.MINUTE, 30); // add 30 minutes to current time // run task every 60 minutes, starting 30 minutes from now 2) java.util.Timer timer = new Timer(); timer.scheduleAtFixedRate( new CacheScourTask(), c.getTime(), (long) 1000 * 60 * 60 );
3) private class CacheScourTask extends java.util.TimerTask {
public void run() {
StringBuffer sbuff = new StringBuffer();
4) DiskCache.cleanCache(100 * 1000 * 1000, sbuff); // 100 Mbytes
sbuff.append("----------------------\n");
5) log.info(sbuff.toString());
}
}
...
// upon exiting
6) timer.cancel();
 This document was last updated on July 2013
This document was last updated on July 2013